Part 1 of this three-part series discussed the need for a simplified cannabis labeling system, such that the abundance of data gathered at the testing lab can make sense to everyone in the cannabis ecosystem. In Part 2, the DiscOmic labeling system was presented. When exploring how the DiscOmic labeling system is able to analyze and organize the information from the cannabis sample, there are two critical components: statistical analysis using artificial intelligence (AI) algorithms and biochemical enzymology. The two systems interact as independent validation of the groupings and other organizing assumptions that the software uses to generate the label. The AI component is used to explore and discover relationships between terpene groups, and the enzymology component is used to verify and validate the statistical patterns being observed by rooting them in scientific hypotheses that explain the patterns that are observed in cannabis data.
AI component
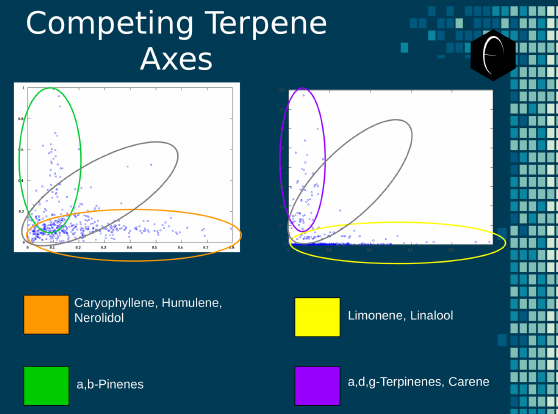
The AI component of the DiscOmic system began with simpler observations rooted in competing terpene axes. A competing terpene axis is when two terpenes, though both commonly at high levels in cannabis, are rarely, if ever, present in high amounts together. This indicates a genetic link, which we will examine in greater detail in the following section addressing enzymology and genetics.
Competing axes, when graphed with one axis representing the concentration of each terpene, visually displays the competition between the terpenes. But this is limited to looking at a graph for each relationship between all of the terpenes, of which 23 were used in our dataset. To look at the correlation of every terpene would mean you would have to generate 253 graphs and analyze each one separately.
In addition to the impracticality of generating 253 graphs for every dataset analyzed, some of the correlations may not be visible when only examining the data in a series of one-to-one relationships. Some patterns are only visible when the sum of one group is compared to another, as is the case with some of the DiscOmic terpene groups. In these scenarios, the amount of terpenes in a group varies widely and seems to have no correlation, but when that group’s data is measured as a sum, very clear patterns start to emerge. We have illustrated two sets of terpene group axes in Figure 5 below. This shows how a grouping selection can illuminate patterns in the data that were not previously accessible and would have been less likely to be discovered with a one-to-one relationship approach.
Figure 5: An illustration of the oppositional nature of competing terpene groups in two scatter diagrams. The left diagram compares the sum of the sesquiterpenes caryophyllene, humulene, and nerolidol to the sum of alpha and beta pinene. The right diagram compares the sum of limonene and linalool with the sum of the terpinenes and carene. All data has been normalized based on the dataset.
An approach that allows for the inclusion of both rapid multi-component comparison and the discovery and identification of stable groupings is an AI technique called self-organizing maps. In this data analytics technique, the data is plotted with every value being an independent axis. Because there can only be three spatial axes, and this model uses 23, the data is grouped by similarity in a higher dimensional space. Once the data is scattered out in hyperspace, a process called unsupervised learning begins to collect patterns in the data. Unsupervised learning refers to the algorithm not being aware of any group labels, and blindly looking for patterns in the hyperspace grouping. The result is that a dataset with 23 variables can be reduced into a dataset with six numbers, without losing significant information. In this way the process can be likened to a data compression program that creates a .zip archive file, because it reduces information in a way that does not lose anything that cannot be reconstructed from the compressed file itself.
To better understand how human and robot minds created the six groups in the DiscOmic label system and why they are ideal for representing the cannabis terpene profile, we will examine some of the self-organizing maps that were created by unsupervised learning algorithms.
Figure 6: A diagram illustrating the concept of a self-organizing map. On the bottom, the variables, x1, x2, etc, each have a connection to a sample represented on planar grid. The result is a heat map with a hot-spot over the cluster of samples representing the clustering of one of the variables.
The concept of a self-organizing map is essentially a plane in a higher dimensional space upon which each variable cluster can be projected. Because a plane is two-dimensional, we can visualize this space, and using colors in a heat map system, we can visualize the diffusion or spread of the clustering as its projection intersects with the plane.
In the case of self-organizing maps describing cannabis samples, each unit on the map represents a sample or group of samples. They are organized in the cluster sharing the feature or grouping of features, in this case high concentrations of certain terpenes. The self-organizing map is set to one terpene; the heat-map shows the cluster of that terpene in the grid. As the self-organizing maps of different terpenes are compared, the clustering within the group of samples can be seen readily, and the terpene groups can easily be identified.
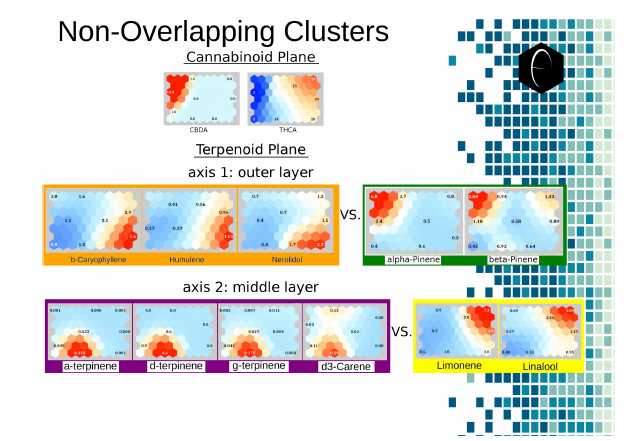
Figure 7: Illustration of two planes of cannabis data illustrated as self-organizing maps. The first plane shows only two components, CBDA and THCA. The second plane shows several terpene components which are grouped into opposing or non-overlapping clusters.
Examining the terpenoid plane across the major terpenes, in Figure 7, shows four major cluster spots, roughly corresponding to the four corners of the plane. The diametric opposition of the orange and green groups and the purple and yellow groups is graphically visible in the plane of the self-organizing maps. With these eleven two-dimensional images, we have been able to correlate the kind of data that would have taken hundreds of two-dimensional images generated with the traditional scatter-graph approach.
Enzymology component
Once the AI component of the analysis generates tightly correlated groupings and matches them with competitive groupings, we have the three concentric layers of the DiscOmic system. At this point we would have groupings that make statistical sense, but we have no underlying biochemical theories to support the observed patterns. To give strength to the organizational scheme, we have created underlying enzymological hypotheses for each of the three rings used in the DiscOmic system. Before we review the relevant data as it relates to enzymology in cannabis, let’s do a quick review of enzymology, biosynthesis, and genetics, and how all three relate.
Figure 8: an overview of how enzymes mediate the reactions that occur during biosynthesis. Clockwise from top-left: A scheme for an anabolic enzyme that builds chemical bonds; a scheme for a catabolic enzyme that destroys chemical bonds; and an example of a biosynthetic enzyme mediated pathway of limonene.
Figure 8 provides a quick overview of the mechanism of enzymes. Enzymes are either anabolic, meaning that they build bigger molecules by forming chemical bonds, or catabolic, meaning they break molecules down by cleaving chemical bonds. Many enzymes have activity of both anabolism and catabolism, but every enzyme must be a member of at least one of the two categories. By using combinations of these enzymes that target different bonds and different molecules, nearly any molecule imaginable can be constructed.
It is these linear biosynthetic pathways that are the link in a plant- or truly any organism- between genetic information and physical traits. If a genome is a library, genes are like books, and each book is a set of instructions on how to build a specific enzyme. Depending on which genes the organism inherited, the enzymes will be physically different. This physical difference in the enzyme is then linked with physical differences in molecular structures made by that enzyme. It is these differences in molecular structure that are the cause of the differences in physical traits in the whole organism. Because each gene is a set of instructions for a specific enzyme, each gene is linked to one enzyme.
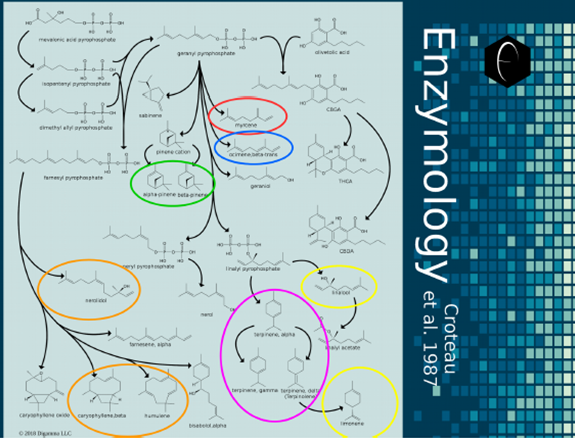
The construction process can be thought of as a long assembly line, where each enzyme makes its bond modification and then passes the molecule to the next enzyme in the line. This is graphically represented as biosynthetic pathways, like the one illustrated above for limonene. Although humans, for simplicity of interpretation, like to represent the pathway to one molecule as a straight line, the truth is that many molecules are made from common precursors, causing our linear biosynthetic pathways to branch out. Inside the cells of the organism, these pathways are all happening in parallel simultaneously, and the precursor reaction must be producing enough precursor to distribute to all the enzymes. If there is not enough precursor to keep every enzyme at full capacity, they then begin competing for available precursors, often with the winners being faster-reaction enzymes and the losers being slower-reaction enzymes. To better illustrate the complexity of terpene biosynthesis and enzymology, we have constructed a biosynthetic mapping of terpenes and cannabinoids based on the research paper by Rodney Croteau published in 1987 on terpene synthesis. [1] Although Croteau’s paper did not perform experiments in cannabis plants, the terpene synthesis pathways examined were similar enough tocreate a biosynthetic landscape within which the enzymological analysis of cannabis could operate.
Figure 9: Map illustrating the enzymology and biosynthesis of terpenes in higher plants. The universal precursor, mevalonic acid pyrophosphate, can be seen the top-left of the image. Arrows indicate enzymatic transformations and branch out in all directions to a variety of terpenes and intermediates. Colors from the DiscOmic system have been used to circle the major terpenes in each terpene group.
The complexity and coordination involved in biosynthesis is evident in the enzymological map. The highlighting of the major terpenes in each of DiscOmic’s terpene group illustrates the complexity of finding these patterns, and why the AI measures described in the previous section were necessary for establishing definite patterns. Because the enzymology of the synthesis of terpenes is so complex, to better understand each DiscOmic terpene group, we will examine a biosynthetic map that focuses exclusively on the compounds of interest and their common precursors.
Figure 10: Enzymology mapping of the relationship between the outer-ring groups 1 and 2. Intermediates illustrated as black circles, and enzymes are indicated in blue. Terpenes are indicated by major group color.
The first map, the sesquiterpenes (orange) vs. the pinenes (green) is a straightforward inverse relationship. The best way to chemically explain inverse statistical relationships is to describe a system with a common precursor and two competing enzymes. In this scheme the common precursor is labeled as precursor 1. P1 is hypothesized to be geranyl pyrophosphate based on the available data. [201] Two enzymes, E1 and E2, compete for this precursor to generate intermediates that can either become pinenes or sesquiterpenes. Additional enzymes may mediate the distribution of terpenes within that group, such as E3 and E4 mediating P2, believed to be farnesyl pyrophosphate based on available evidence.[2]
Enzymological models such as this one can easily explain the phenomenon, covered in the AI section, where individual terpenes fail to have any correlation but show strong correlation in a group. This would explain how the total of the group would be a more consistent number than the components, because the components of that group are all derived from a common precursor.
Figure 11: Enzymology mapping of the relationship between the middle-ring groups 4 and 3. Intermediates illustrated as black circles, and enzymes are indicated in blue. Terpenes areindicated by major group color.
The second map, the terpinenes (purple) vs. the limonene-linalool (yellow) is also a straightforward inverse relationship. Again, we hope to describe a system with a common precursor and two competing enzymes to explain the observed correlation. In this scheme (Figure 11), the common precursor is labeled as precursor 1. P1 is hypothesized to be linalyl pyrophosphate based on the available data. [1] Two enzymes, E1 and E2, compete for this precursor to generate intermediates that can either become terpinenes or limonene-linalool. Additional enzymes may mediate the distribution of terpenes within that group, such as E3 and E4 mediating P2, believed to be alpha-terpinyl pyrophosphate.[1]
Once again, we see a scheme where individual terpenes fail to have much correlation but illustrate correlation as a group. The reasoning is based on the stable number of the total, contrasted with the more chaotic competing enzymes that do the “final touches” on terpenes synthesis.
Figure 12: Enzymology mapping of the relationship between the inner-ring groups 6 and 5. Intermediates illustrated as black circles, and enzymes are indicated in blue. Terpenes are indicated by major group color.
The third map, ocimene (blue) vs. myrcene (red) is also a more complex relationship than was observed in the first or second map. Ocimene and myrcene are neither inversely correlated, where the presence of one competes with the other, nor directly correlated, where the amount of one is tied to the amount of the other in a fixed ratio. This relationship shows no observable correlation, yet both terpenes are present at such high concentrations that they constitute a substantial portion of a cannabis sample’s terpene content. The lack of correlation between the two cannot be explained very easily, because both are known to be made through precursor geranyl pyrophosphate. [1]
The best theory that fits the current evidence is that both ocimene and myrcene can be made from a common precursor, but each terpene can also me made from derivatives of that precursor.[1] If ocimene and myrcene are made from identical precursors, we would see them in a fixed ratio. But, if ocimene and myrcene are able to interact with different derivatives of geranyl pyrophosphate, then it would allow two independent numbers to be derived from the same baseline value of precursor. What would influence the final number of these two terpenes would be a mixture of enzymes, some affecting the myrcene pathway, some the ocimene pathway, and many having distinct effects on both.
This degree of complexity is very common in enzymological schemes, especially in higher plants such as cannabis. With complications such as enzyme promiscuity, a phenomenon where one enzyme can actually cross-catalyze different reactions at different rates, these patterns can become even more challenging to understand.[3] It is for this reason that tools for the consuming public, the cannabis industry, and the cannabis research community, are needed to help humans begin to benefit from the complexities of cannabis, even as the final aspects of our scientific understanding of the cannabis and human organisms are still being discovered.
References
[1] Croteau, Rodney. “Biosynthesis and catabolism of monoterpenoids.” Chemical Reviews 87.5 (1987): 929-954. [2] May, Bianca, B. Markus Lange, and Matthias Wüst. “Biosynthesis of sesquiterpenes in grape berry exocarp of Vitisvinifera L.: evidence for a transport of farnesyl diphosphate precursors from plastids to the cytosol.” Phytochemistry 95 (2013). [3] Hult, Karl, and Per Berglund. “Enzyme promiscuity: mechanism and applications.” Trends in biotechnology 25.5 (2007): 231-238.