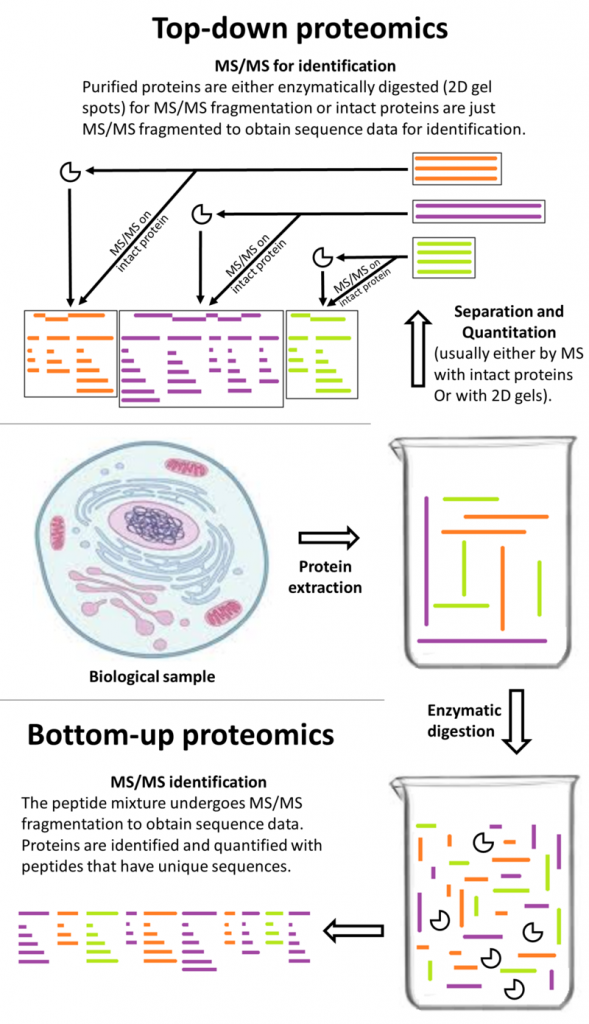
Proteomics is the study of an organism’s proteins, including protein composition, function, interactions, etc. It is considered more complicated than genomics due to constantly fluctuating conditions but conveys a clearer picture of an organism’s actual functioning. Contributions to the proteomics of medical cannabis took flight in 2019 with three studies authored by Vincent et al. [1-3]
The first study exposed mature cannabis flower to six different protein extraction methods. The best method—that which allowed identification of 435 peptides, mostly involved in secondary metabolism (e.g., phytocannabinoid synthesis)—was a trichloroacetic acid/acetone precipitation step and a guanidine hydrochloride (guanidine-HCl) buffer. The authors declared it the “first time protein extraction is optimised from cannabis reproductive organs,” noting “the guanidine-HCl buffer employed…has never been used before on C. sativa samples.” [1] This is bottom-up, or shotgun, proteomics (see figure).
Stuart Christopher Brown, CC By-SA 4.0
The next study [2] tested several top-down proteomics methods (using liquid chromatography-mass spectrometry, LC-MS) to identify small to medium, intact cannabis proteins (rather than digested peptides). The researchers were able to label 46 proteins, almost all of them connected to primary metabolism (e.g., photosynthesis). In this case, the research group admitted that “[d]ata analysis proved challenging, and most of the MS/MS spectra remained unannotated for a reason yet to be elucidated.” [2]
The third and final investigation published by Vincent et al in 2019 sought to examine the issue from a different angle, namely the best way to digest medicinal cannabis proteins intended for bottom-up and middle-down analyses. [3] Middle-down proteomics uses the same procedure as the bottom-up strategy but aims for larger peptides that may otherwise be missed. [4] To accomplish this, the key component is the protease, or enzyme that breaks down (digests) the protein into peptides. [4]
The research team used trypsin in their first study as the “gold standard” protease for the digestion step. [1,3] In the most recent study, however, they played with a few other options for digestion with the goal of maximizing peptide yields:
- Mixture of trypsin and endoproteinase LysC
- Endoproteinase GluC
- Chymotrypsin
The effects of these proteases were investigated in three ways: individually (e.g., trypsin/LysC), sequentially (trypsin/LysC then GluC), and together (trypsin/LysC and GluC).
The proteases behaved uniquely and demonstrated distinct results. Combining the proteases led to the detection of over 27,000 mature cannabis peptides, including longer peptides attributed to GluC. This amounts to 229 distinct proteins. The researchers outlined their respective functions, roughly, as follows:
- Primary metabolism: 31%
- Secondary metabolism: 23%
- Gene expression: 19%
- Hormones: 14%
- Unknown: 10%
Importantly for cannabis geneticists and breeders, the authors concluded they had captured “all the phytocannabinoid-related enzymes and most of the enzymes involved in terpenoid backbone biosynthesis.” [3] This included, for example, (+)-alpha-pinene synthase and a novel cannabidiolic acid-like synthase. Multiple proteases in the digestion step therefore provided a major advantage. Ultimately, the research aims to “validate the annotations of genome sequencing projects in a proteogenomic manner and facilitate breeding programs.” [3]
References
- Vincent D, et al. “Optimisation of Protein Extraction from Medicinal Cannabis Mature Buds for Bottom-Up Proteomics.” Molecules, vol.24, no.4, 2019, p. 659. Impact Factor: 3.060; Times Cited:11
- Vincent D, et al. “Top-Down Proteomics of Medicinal Cannabis.” Proteomes,7, no.4, 2019, p.33, doi:10.3390/proteomes7040033. Impact Factor: 2.770; Times Cited:3
- Vincent D, et al. “A Multiple Protease Strategy to Optimise the Shotgun Proteomics of Mature Medicinal Cannabis Buds.” Int J Mol Sci, vol.20, no.22, 2019, p.5630. Impact Factor: 4.183; Times Cited:1
- Cristobal A, et al. “Toward an Optimized Workflow for Middle-Down Proteomics.” Analytical Chemistry,89, no.6, 2017, pp.3318-3325, doi:10.1021/acs.analchem.6b03756. Impact Factor: 6.350; Times Cited:30
Images:
Wang S., Trumble WR, Liao H, Wesson CR, Dunker AK, Kang C, and Astrojan, Wikimedia Commons, CC-By 3.0